AVDENS_GF0906 - calculates the average density of gf0906
Author: Torsten Linders
Date: 7 June 2010
Contents
Preparations
Cleaning up
clear
close all
Loading variables from mat-files
load latlon dist_c load epsi5dm dens load mycolormap mycolormap
Plotting parameters
casts=size(dens,2); %Number of casts, profiler data. imax=45; %Number of columns in transect. hmax=9e3; %Horizontal distance in transect. hfact=hmax/imax; %Metres represented by each column. jmax=size(dens,1); %Number of rows in transect. dmax=120; %Maximum depth in transect. dfact=dmax/jmax; %Metres represented by each row.
Allocations
densAv=zeros(jmax,imax); %Allocating matrix, mean density. densAv_count=zeros(jmax,imax); %Allocating matrix, number of obs. densAv_v=zeros(jmax,1); %Allocating vector, mean density. densAv_vcount=zeros(jmax,1); %Allocating vector, number of obs. f=zeros(1,casts);
Transect
for i=1:casts i_m=round(dist_c(i)/hfact); if i_m>0 for j=1:jmax if dens(j,i)>0 densAv(j,i_m)=densAv(j,i_m)+dens(j,i); densAv_count(j,i_m)=densAv_count(j,i_m)+1; end end end end densAv=densAv./densAv_count;
Profile
for i=1:imax for j=1:jmax if not(isnan(densAv(j,i))) densAv_v(j)=densAv_v(j)+densAv(j,i); densAv_vcount(j)=densAv_vcount(j)+1; end end end densAv_v=densAv_v./densAv_vcount;
The array sigma_v
sigma_v contains values for isolines. It is used for the contour plots of density (in legs_gf0906). The purpose of these plots is to show structures in the stratification. sigma_v is therefore constructed with the average profile densAv_v as the base. A minimum resolution is applied in order to not get to many contour lines at large depths. sigma_vs is a an array with fewer values, wich can be used for emphasising some contour lines.
The array sigma_j contains the depth indices of the sigma_v values. It is used in subplot 2 below for indicating the depths of the values of sigma_v. sigma_js contains the depth indices of sigma_vs.
sigma_v=zeros(length(densAv_v),1); for i=2:length(densAv_v) sigma_v(i)=max(sigma_v(i-1)+3.0e-2,densAv_v(i)); end sigma_v=sigma_v(7:215); %Indices without NaN. sigma_v=sigma_v(1:105); %Problem with high indices?! sigma_vs=sigma_v(10:10:105); sigma_j=interp1(densAv_v(7:215),7:215,sigma_v); sigma_js=interp1(densAv_v(7:215),7:215,sigma_vs); save sigma sigma_v sigma_vs
Plots
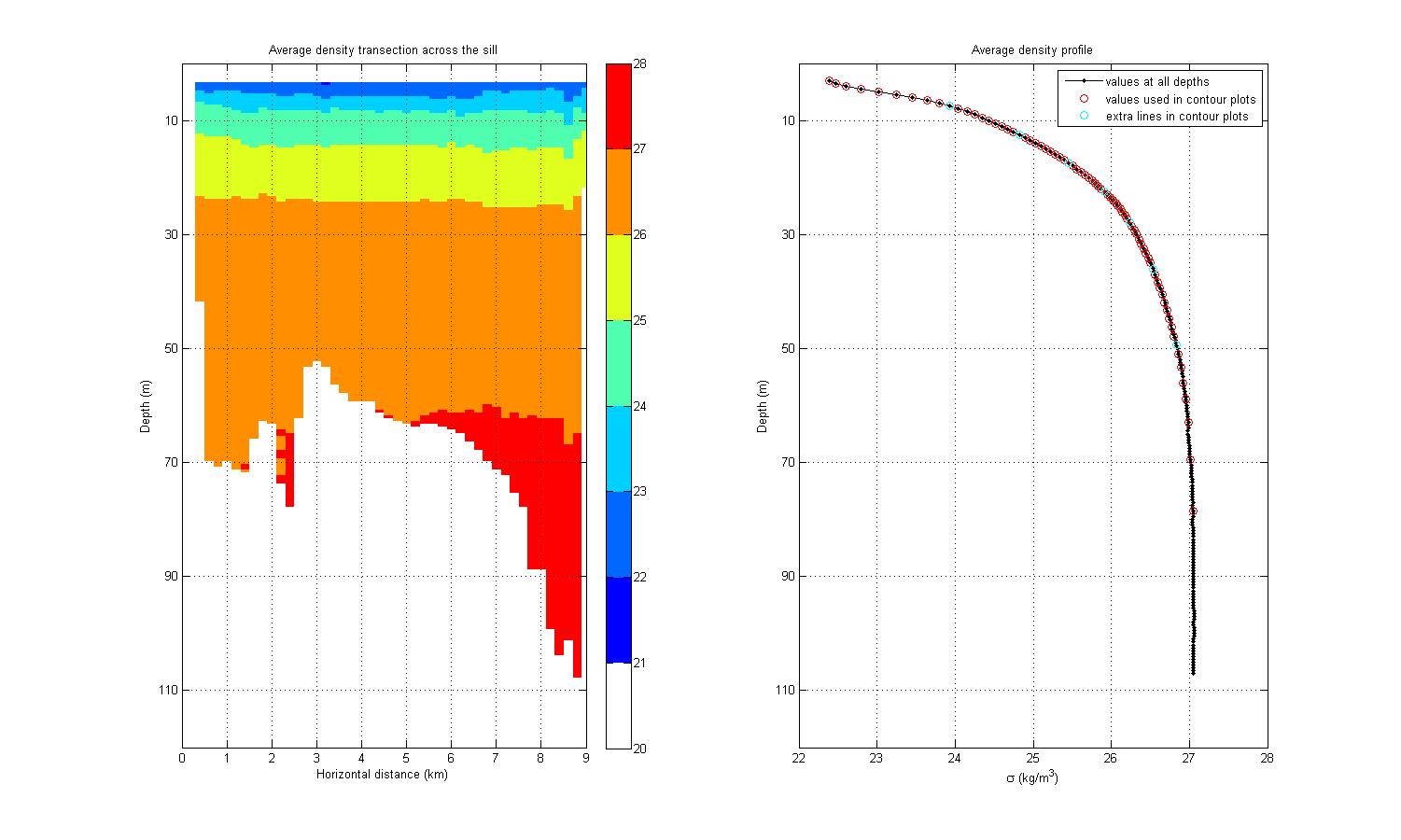
fig=figure(1000); set(fig,'Position',[1 1 1500 900]); set(fig,'Colormap',mycolormap([1 2:9:end],:)); subplot(1,2,1) imagesc(densAv) title('Average density transection across the sill') caxis([20 28]) grid on xlim([0 imax]) ylim([0 jmax]) colorbar xlabel('Horizontal distance (km)') set(gca,'XTick',0:1000/hfact:imax) set(gca,'XTickLabel',(0:1000/hfact:imax)*hfact/1000) ylabel('Depth (m)') set(gca,'YTick',20:20/dfact:jmax) set(gca,'YTickLabel',20*dfact:20:jmax*dfact) subplot(1,2,2) plot(flipud(densAv_v),1:jmax,'k.-',... sigma_v,241-sigma_j,'ro',... sigma_vs,241-sigma_js,'co') legend('values at all depths',... 'values used in contour plots','extra lines in contour plots') title('Average density profile') grid on ylim([0 jmax]) xlabel('\sigma (kg/m^3)') ylabel('Depth (m)') set(gca,'YTick',20:20/dfact:jmax) set(gca,'YTickLabel',jmax*dfact-10:-20:20*dfact)
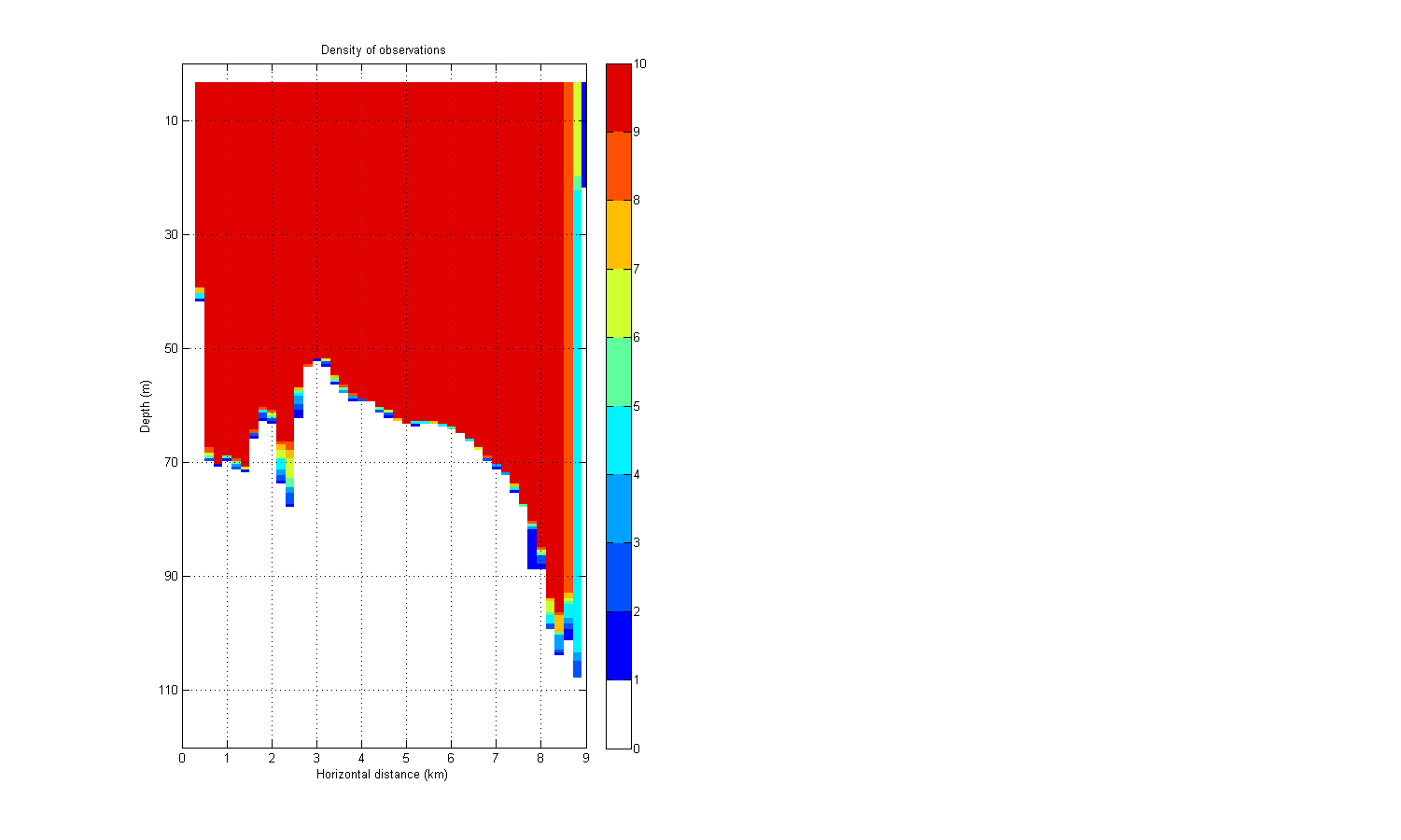
Density of observations
To get a sufficient density of observations in all columns has the horizontal resolution been lowered to hfact=200 m. This governed by the number of columns, imax=45.
if 1 fig=figure(1001); set(fig,'Position',[1 1 1500 900]); set(fig,'Colormap',mycolormap([1 2:7:end],:)); subplot(1,2,1) imagesc(densAv_count) title('Density of observations') caxis([0 10]) grid on xlim([0 imax]) ylim([0 jmax]) colorbar xlabel('Horizontal distance (km)') set(gca,'XTick',0:1000/hfact:imax) set(gca,'XTickLabel',(0:1000/hfact:imax)*hfact/1000) ylabel('Depth (m)') set(gca,'YTick',20:20/dfact:jmax) set(gca,'YTickLabel',(20*dfact:20:jmax*dfact)) end